1 Search for scRNA-seq cell annotation prior data in CellSTAR
1.1 Quick Search for comprehensive annotation prior data
The Quick Search bar on Home page allows you to quickly retrieve scRNA-seq cell annotation prior data based on specific keyword(s).
(1) Enter keyword(s) and perform quick search: To initiate a quick search, enter one or multiple keyword(s) related to your research (separated by space). For example, you can use keywords such as "mouse" (Species); "lung" (Tissue); "Macaca fascicularis colon" (Species & Tissue); "10x Genomics" (Sequencing Technology); "T cell" (Cell Type), etc. Once you have entered the keywords, click on the Search button to perform the search.

(2) Retrieval of results: The search results ideally include two types of relevant prior data, including annotated reference data and canonical marker data (or contain only one type of data). Detailed information on these two types of prior data could be found in sections of "1.2 Search for annotated reference data", "2 Details of annotated reference data of each experiment", "1.3 Search for canonical marker data", and "3 Details of related canonical marker(s) and reference data of each cell type".
1.2 Search for annotated reference data
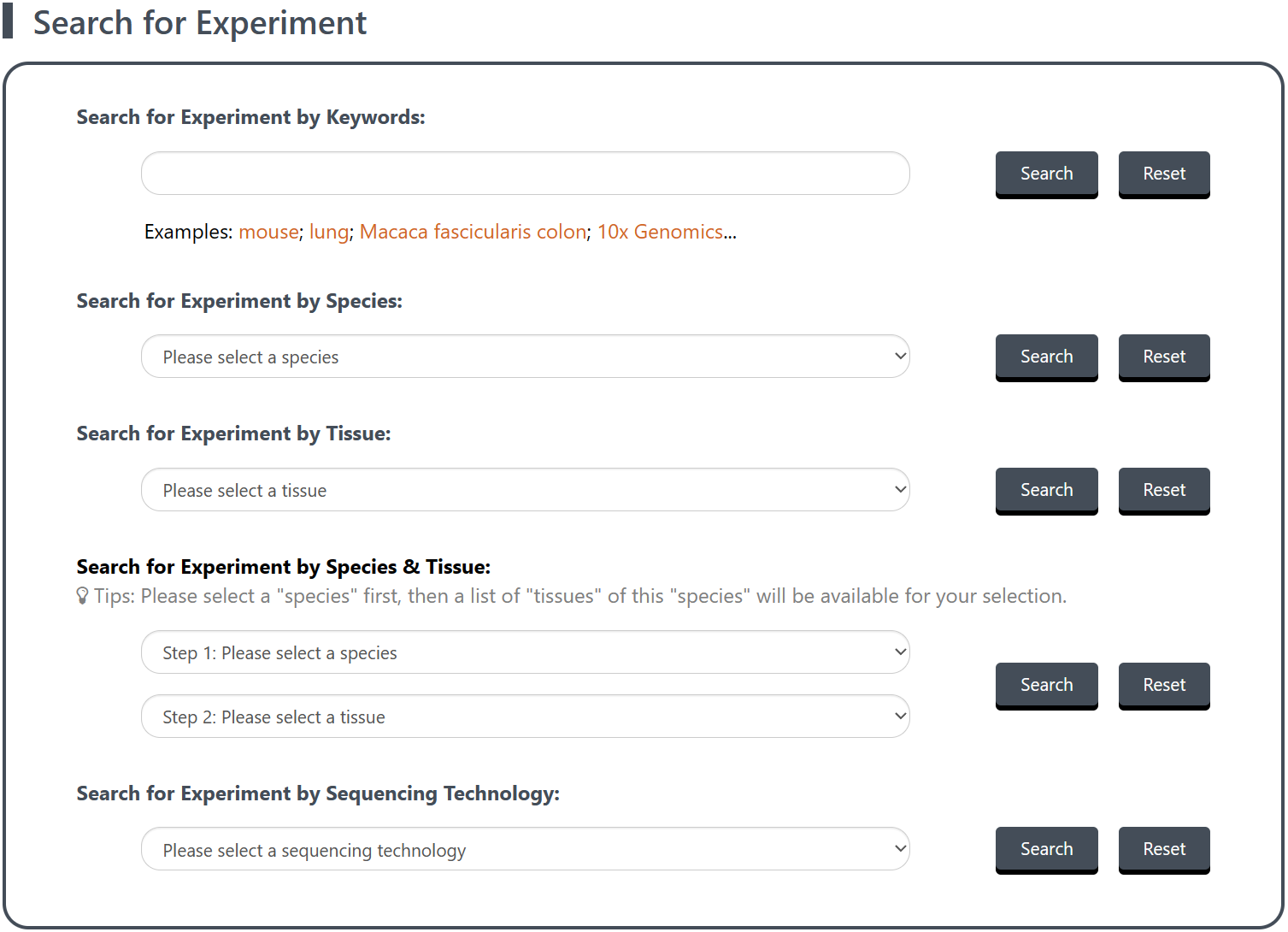
The dropdown menu of Search for Experiment inside the navigation bar of Search allows you to retrieve reference data (i.e., existing expertly annotated single-cell references) associated with scRNA-seq experiment(s).
(1) Select search mode and perform the search: On the Search for Experiment page, five modes are available to provide more accurate and personalized research results, including by Keyword(s), Species, Tissue, Species & Tissue and Sequencing Technology. Once you have determined what to search for, click on the Search button to perform the search.
Note: For all search modes except the keyword search, you can simply select the relevant candidate from the dropdown box.
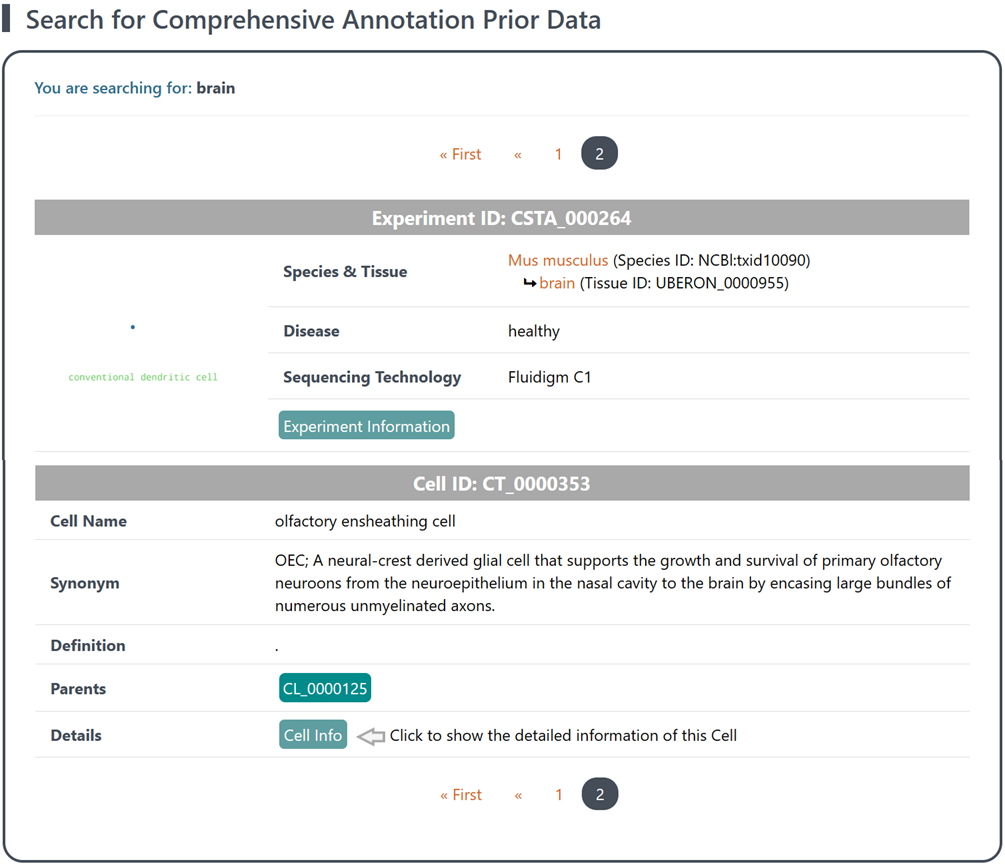
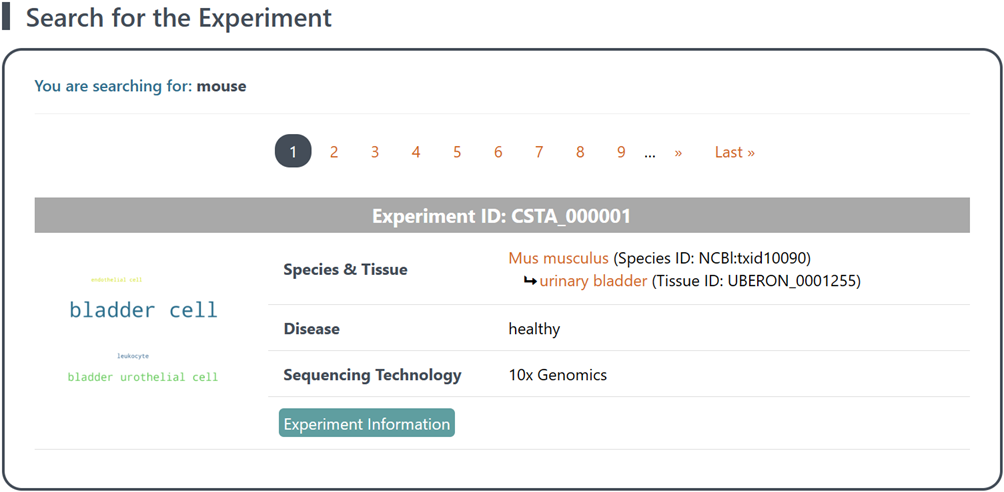
(2) Retrieval of results: The search results will display a list of scRNA-seq experiments (each assigned a unique ID for reference) with corresponding reference data. On the left side of each experiment item, you will find a word cloud map displaying the abundance of distinct cell populations within the reference data, which vividly illustrates the intricate cellular landscape and highlights the prevalence of diverse populations in the dataset. On the right side of each experiment item, you will find general information of the reference data, including the Species, Tissue, studied Disease and the Sequencing Technology used.
(3) Explore metadata and reference data: Click the Exp Info button to access detailed metadata and explore reference data of each experiment item. See section "2 Details of annotated reference data of each experiment" for details.
1.3 Search for canonical marker data
The dropdown menu of Search for Cell Type inside the navigation bar of Search allows you to retrieve canonical marker data (i.e., existing expertly annotated single-cell references) & related scRNA-seq experiments of specific cell type.
(1) Select search mode and perform the search: On the Search for Cell Type page, two modes are available to provide more accurate and personalized research results, including by Keyword(s) and Cell Name. Once you have determined what to search for, click on the Search button to perform the search.
Note: For all search modes except the keyword search, you can simply select the relevant candidate from the dropdown box.

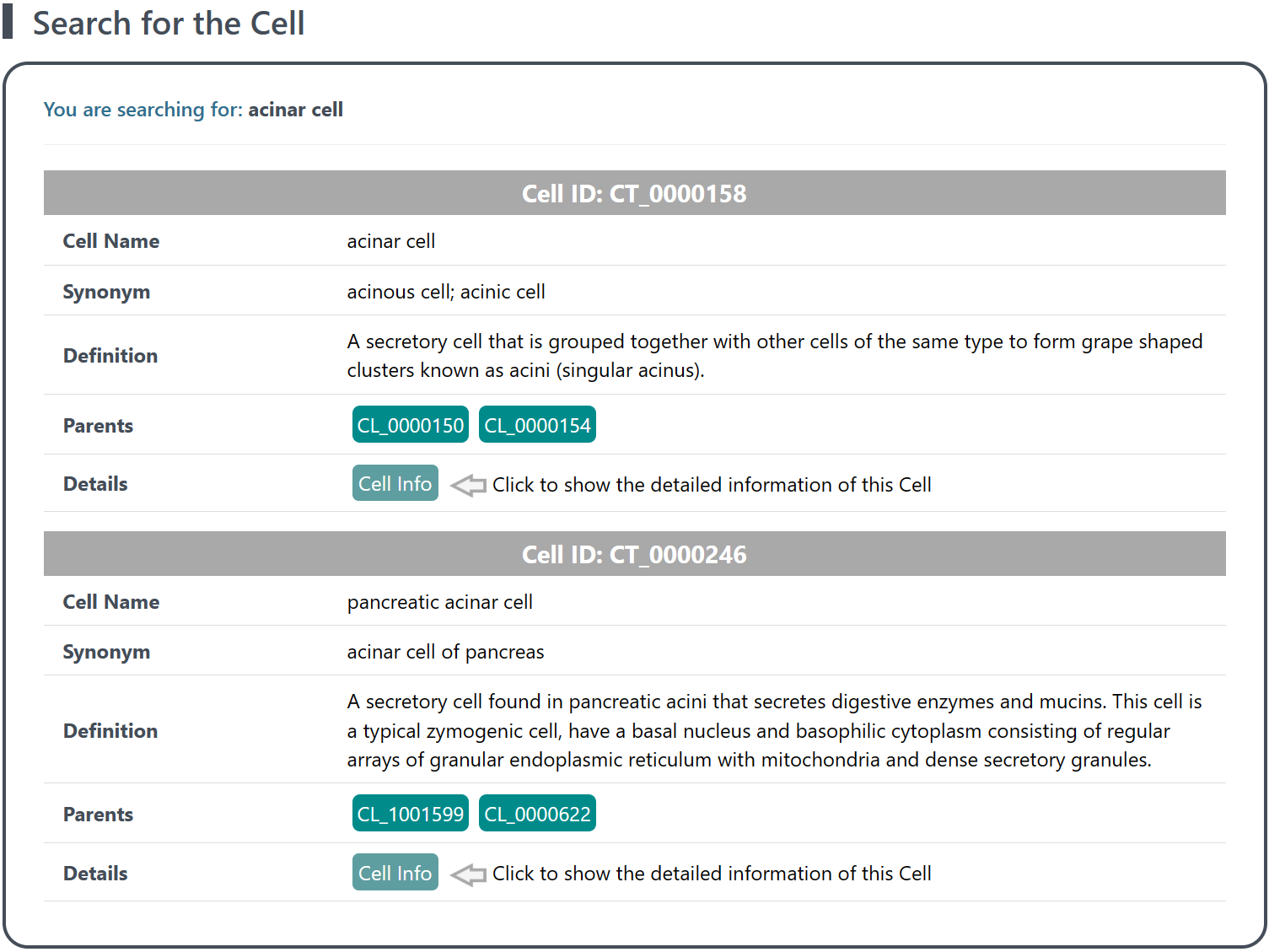
(2) Retrieval of results: The search results will display a list of cell types (each assigned a unique Cell ID for reference). Under each cell item, you will find general information of the cell, including the Cell Name (Synonym), Definition (description of a specific cell type, including morphology, function, location, and other characteristics) and Parents (direct superiors or higher-level cell types of a specific cell type, linking to Cell Ontology Lookup Service of EMBL-EBI), which enables a better understating on the features and hierarchical relationship of different cell types.
(3) Explore related canonical marker(s) and reference data: Click the Cell Info button to access detailed canonical marker(s) and reference data related to each cell type. See section "3 Details of related canonical marker(s) and reference data of each cell type" for details.
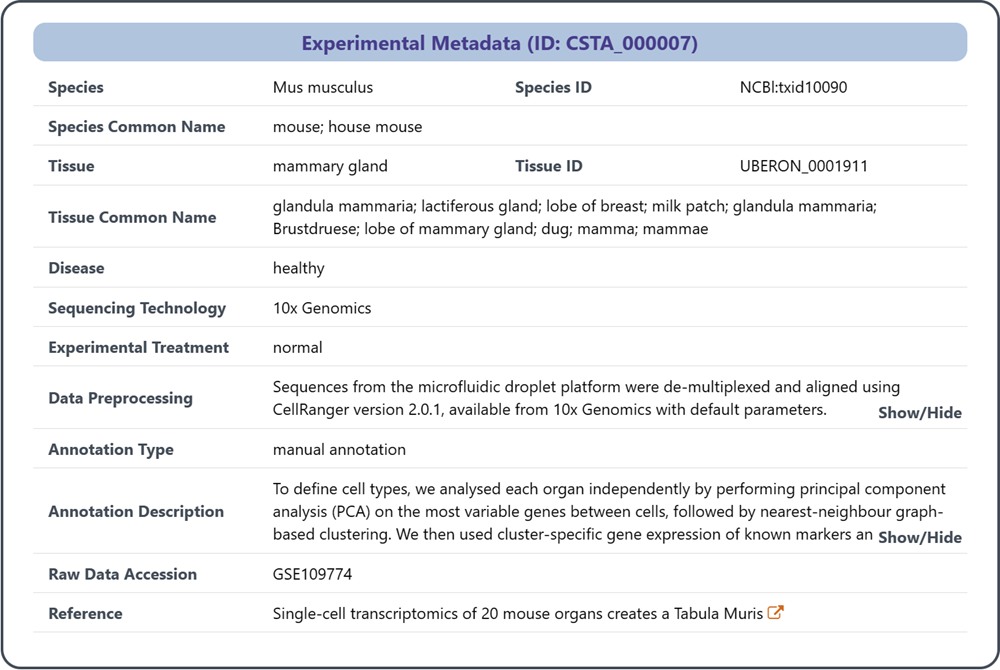
2 Details of annotated reference data of each experiment
This section accurately describes the collection of relevant metadata about the scRNA-seq experiment and its associated reference data, which conveys key context and background information for the experiment.
(1) Species: the unified species name based on NCBI Taxonomy
(2) Species ID: the unique identifier assigned to the species (NCBI Taxonomy)
(3) Species Common Name
(4) Tissue: the unified tissue name based on Uberon
(5) Tissue ID: the unique identifier assigned to the tissue (Uberon)
(6) Tissue Common Name
(7) Disease: the disease studied in the experiment (if applicable)
(8) Sequencing Technology: the sequencing technology used for providing the expression profiles of individual cells
(9) Experimental Treatment: any specific treatment or condition applied during the experiment
(10) Data Preprocessing: details about the preprocessing steps performed on the raw data
(11) Annotation Type: the strategy to label cell types, including manual annotation, automatic annotation, celluar sorting and immunopanning
(12) Annotation Description: additional information about the annotation process
(13) Raw Data Accession: the unique identifier or accession number assigned to the raw data in public repositories
(13) Reference: relevant literature or publication associated with the experimental study
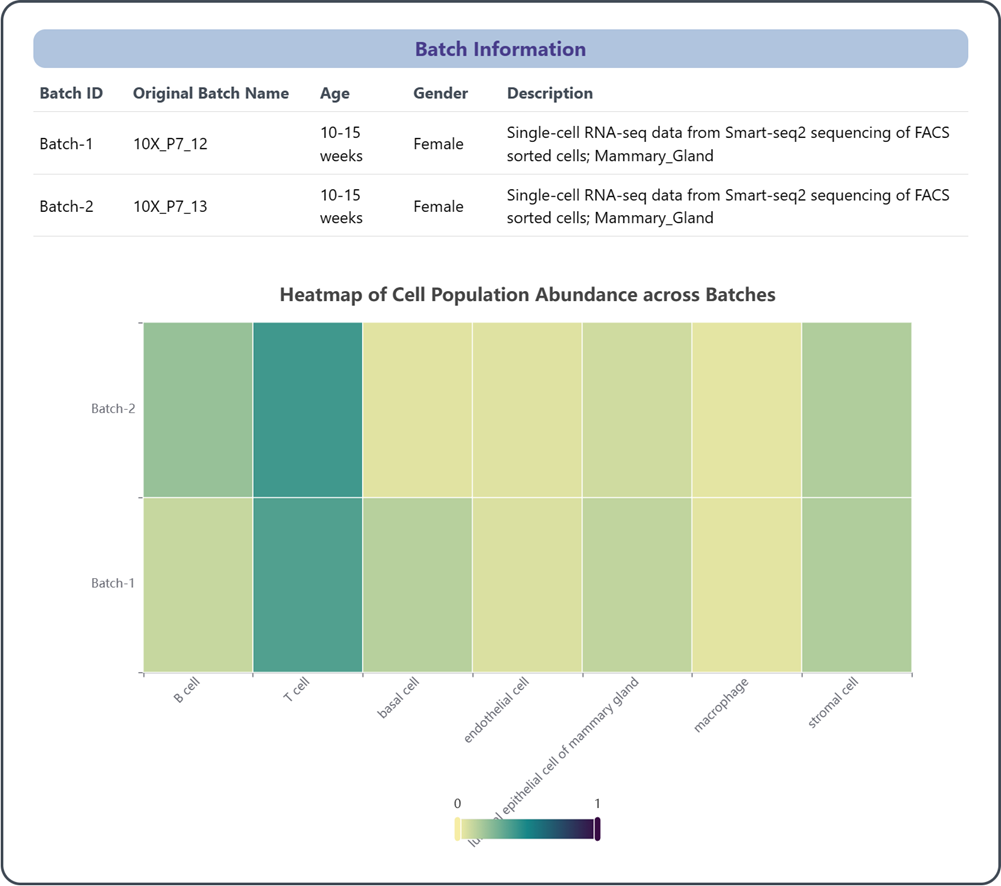
2.2 Batch information
This section provides insights into the characteristics and compositions of different batches of the experiment and reference data, which contributes to understanding the effect of variations in experimental conditions and samples on cell type composition. These information is valuable for identifying potential batch effects, understanding the impact of batch-related factors on the data, and considering appropriate analytical approaches when utilizing the reference data.
(1) Batch ID: the unique identifier assigned to the batch
(2) Original Batch Name: the original name or label associated with the batch
(3) Age, Gender, Description: age, gender and other relevant descriptions on samples included in the batch
(4) Heatmap of Cell Population Abundance across Batches: This heatmap enables users to visually analyze and compare the distribution and variability of cell populations among different batches
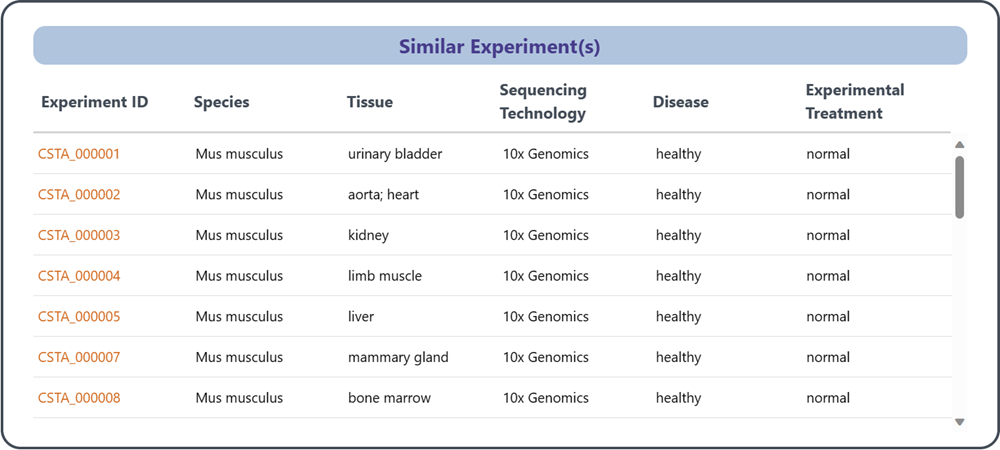
2.3 Similar Experiments
This section establishes connections among datasets associated with the same literature or common research project, which facilitates navigation through related datasets while demonstrating consistency and comparability in metadata across different experiments.
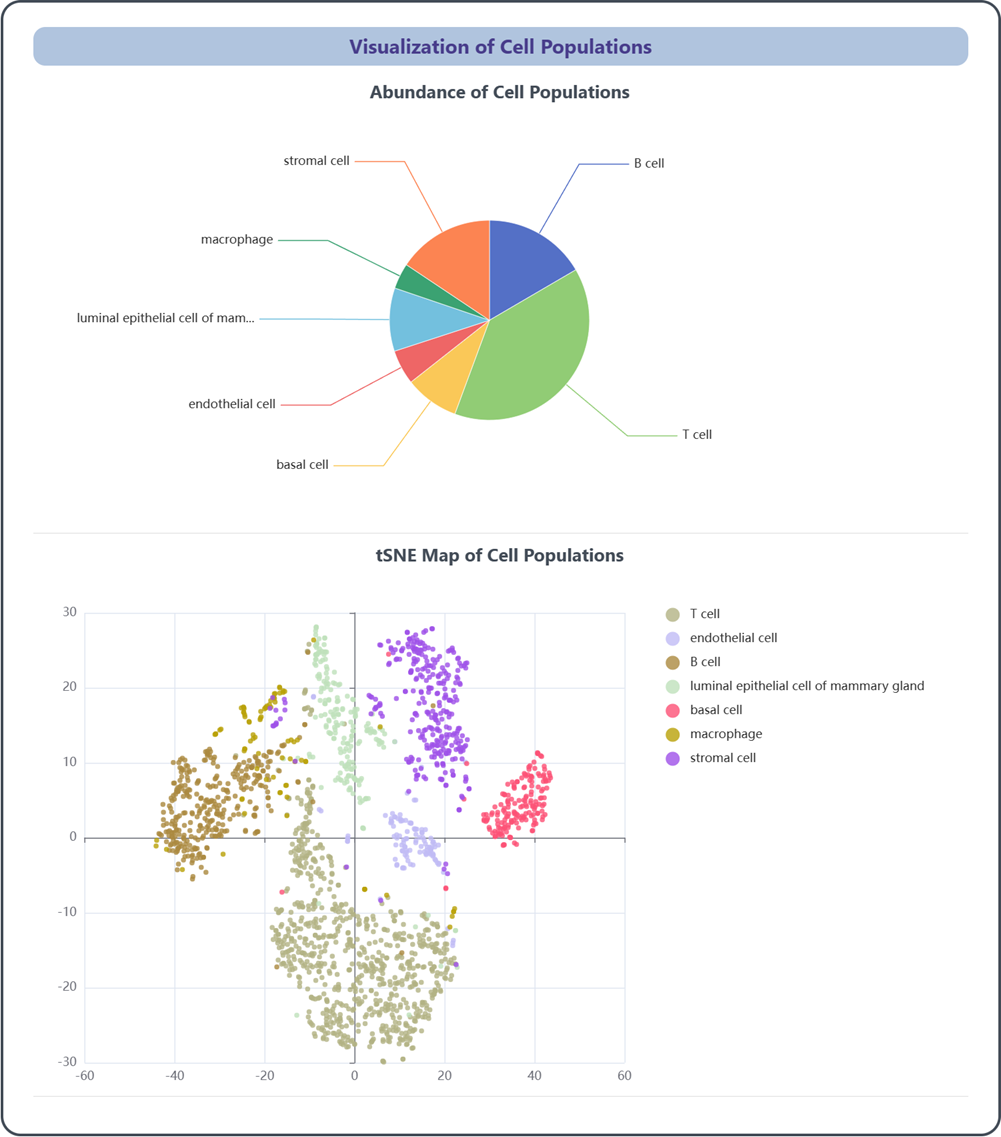
2.4 Visualization of cell populations
This section provides visualizations of various cell populations within the reference data. Each figure can be downloaded by clicking the Save as Image button.
(1) Abundance of Cell Populations: This pie chart provides an overview of relative abundance of different cell populations within the reference data, giving users a quick understanding of the distribution and composition of cell populations.
(2) tSNE Map of Cell Populations: This tSNE map displays the intricate spatial relationships of diverse clusters based on their gene expression profiles.
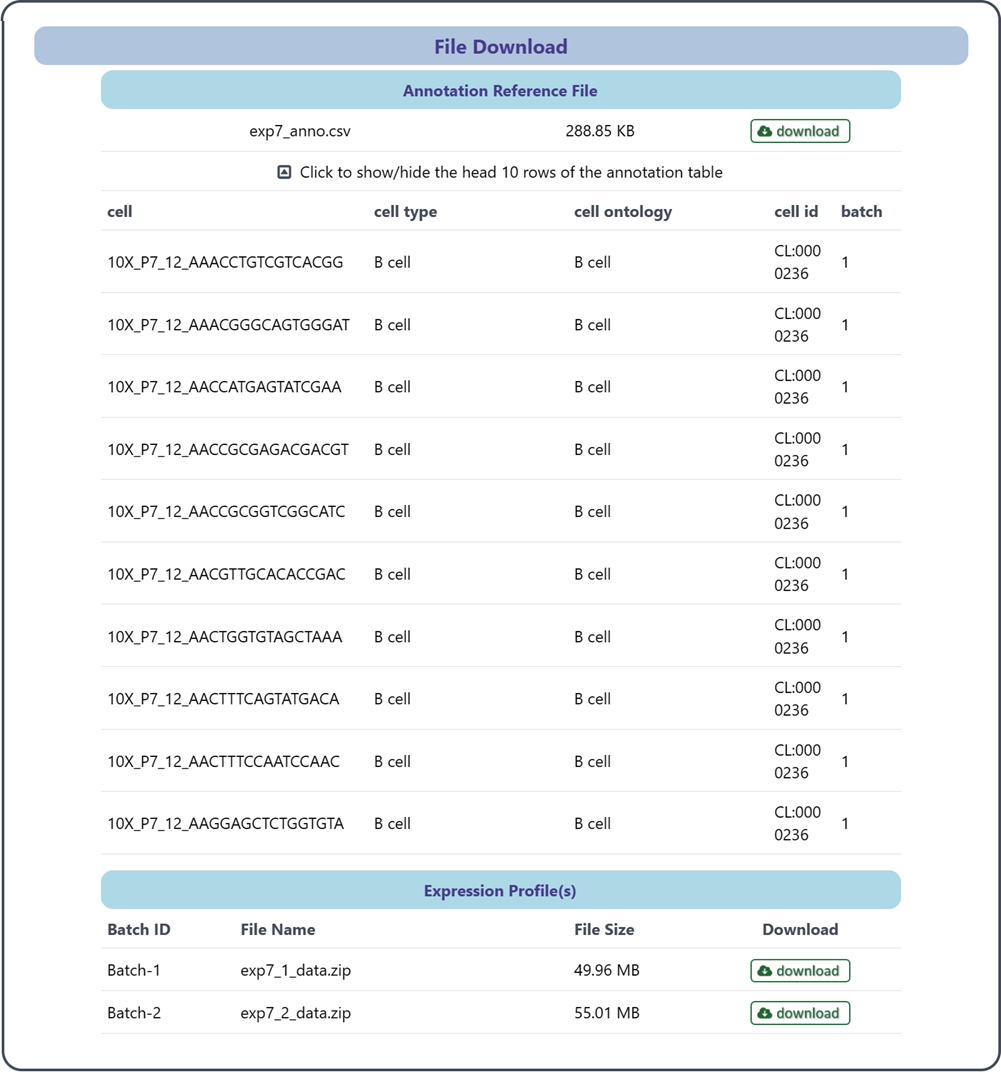
2.5 File download
This section allows you to access and download reference data which include two types of files: an annotation reference file and expression profiling file(s). Select the file you want and click the Download button, choose a destination on your local system and save the file.
(1) Annotation reference file: This file provides a mapping between cells and their corresponding annotated cell types for all batched in the expression profiling file(s). This file serves as a crucial resource for associating specific cell identities with their corresponding cell type annotations, which can be utilized to enhance your analysis, annotation, or classification tasks.
(2) Expression profiling file(s): The expression profiling file(s) contain quantitative gene expression levels across cells, which enable users to explore gene expression patterns, identify differentially expressed genes.
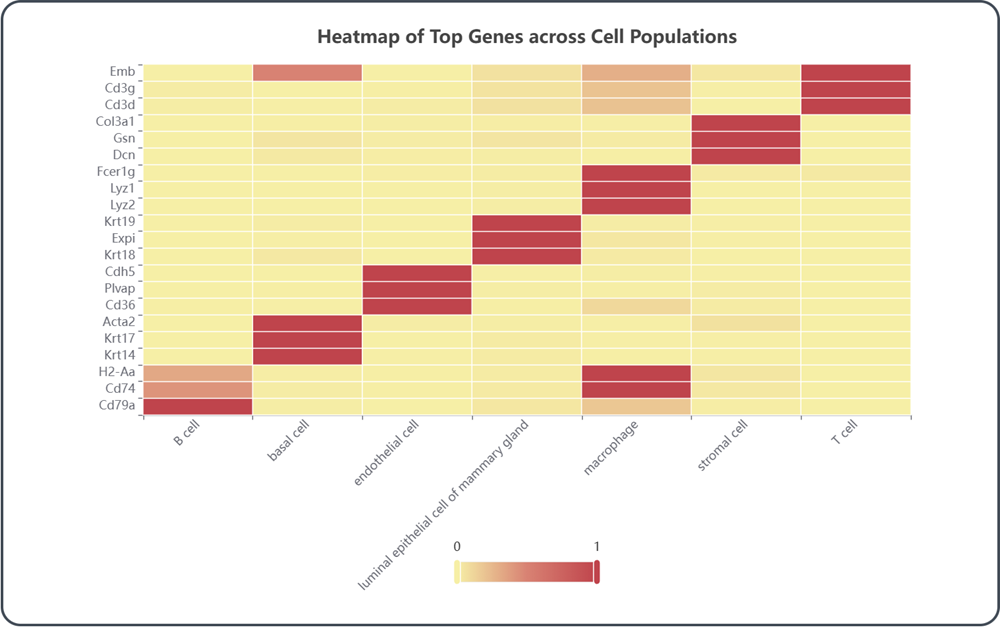
2.6 Visualization of top genes
Heatmap of Top Genes across Cell Populations: This heatmap represents the expression patterns of top differentially expressed genes across different cell populations, which facilities the identification of potential molecular drivers underlying complex biological processes.
3 Details of related canonical marker(s) and reference data of each cell type
On the page of Details of Cell Information, detailed information on canonical marker(s) and reference data related to each cell type is provided, including general information of the cell, cell-related experiment(s), and cell-related canonical markser(s).
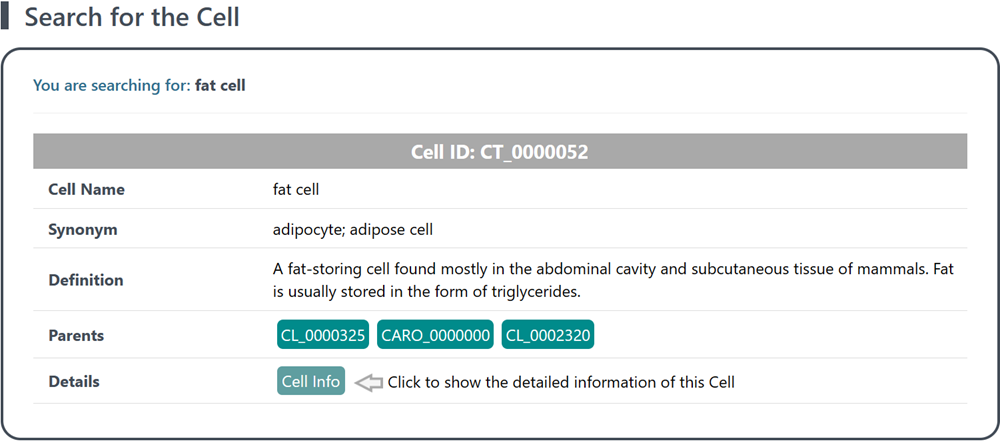
3.1 General information of the cell
This section provides the identity, morphology, function, location, and other characteristics of the cell, and enables a better understating on the lineage relationships or hierarchical structures between different cell types. These information is valuable for the identification and characterization of specific cell type, and the exploration between cell types. Additionally, the external link provides a convenient way to access further information or external resources associated with the cell.
(1) Cell: the unified cell name based on Cell Ontology
(2) Cell ID: the unique identifier assigned to the cell (Cell Ontology)
(3) Synonym: any alternative names associated with the cell
(4) Definition: a brief description of the cell, including morphology, function, location, and other characteristics
(5) Parents: the direct superiors or higher-level cell types of the cell type, linking to Cell Ontology Lookup Service of EMBL-EBI
(6) External Link: the URLs linking to Ontobee and the Cell Ontology Lookup Service of EMBL-EBI
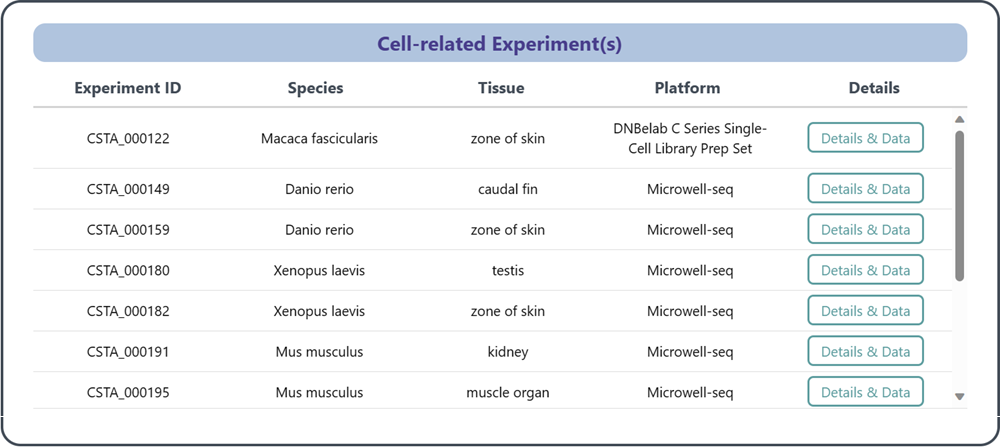
3.2 Cell-related Experiment(s)
This section provides a list of reference data associated with scRNA-seq experiment(s), which contain the specific cell type. In addition to the general experimental metadata, the URL linking to the section "2 Details of annotated reference data of each experiment" is provided for details.
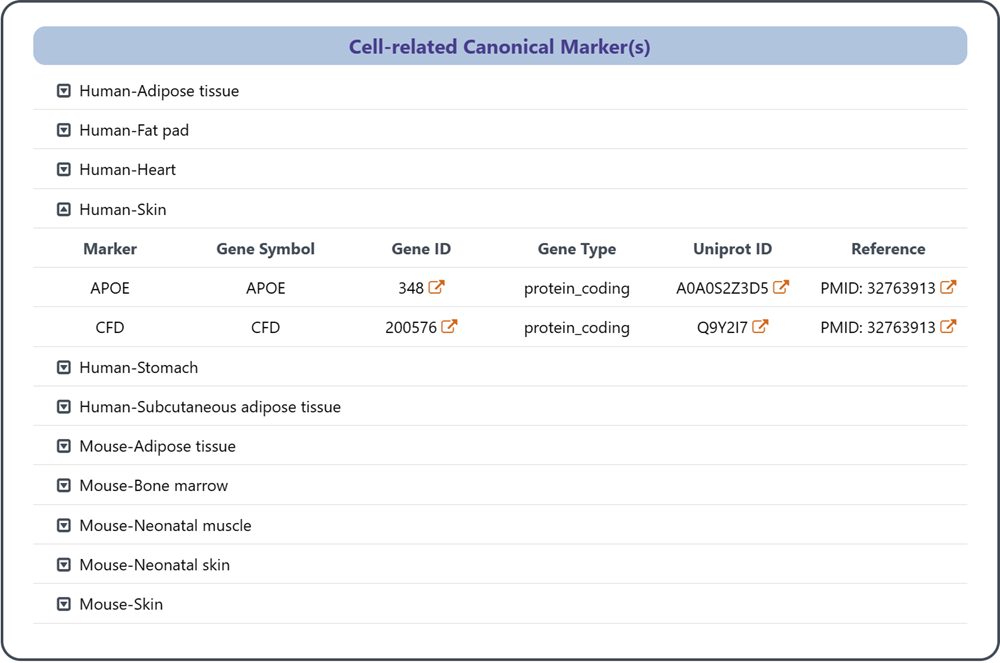
3.3 Cell-related Canonical Markser(s)
This section provides a list of canonical markers associated with the specific cell in different tissues.
(1) Marker: the unified marker name based on Entrez Gene
(2) Gene Symbol: the unique gene symbol assigned to the gene marker (Entrez Gene)
(3) Gene ID: the unique identifier assigned to the gene marker (NCBI Gene)
(4) Gene Type: the category of the gene marker (NCBI Gene)
(5) Uniprot ID: the unique identifier assigned to the protein encoded by the gene marker (Uniprot)
(6) Reference: the URL linking to the publication in PubMed
4 Browse annotated reference data in CellSTAR
The browse function in CellSTAR helps you to streamline the exploration of reference data and facilitates the discovery of experiments relevant to a particular species/tissue.
4.1 Browse annotated reference data by species
The dropdown menu of Browse by Species inside the navigation bar of Browse allows you to browse all available reference data in CellSTAR by species.
(1) Browse species entries: Users can quickly scan through the list of species entries (sorted by number of experiments) to identify the species of interest. Each species entry provides basic information about the species, along with the number of scRNA-seq experiments associated with it.
(2) Access reference data of specific species: Locate the desired species, and click the Search button provided within the species entry. It will direct you to the Search for Experiment page to access reference data. See section "1.2 Search for annotated reference data" for details.

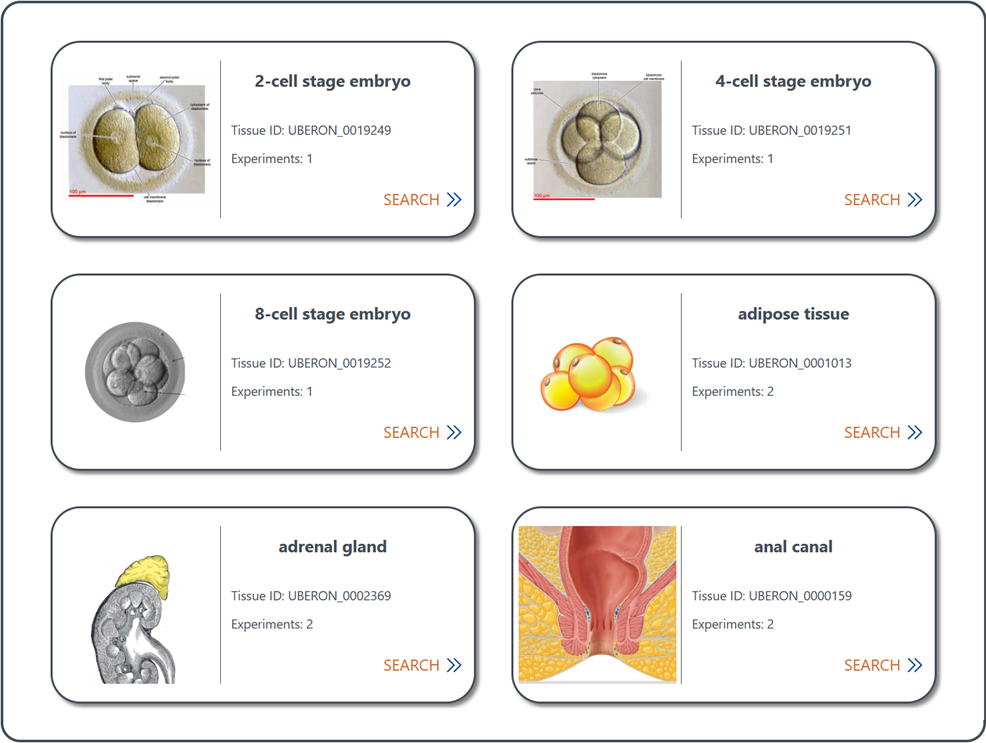
4.2 Browse annotated reference data by tissue
The dropdown menu of Browse by Tissue inside the navigation bar of Browse allows you to browse all available reference data in CellSTAR by tissue.
(1) Browse tissue entries: Users can quickly scan through the list of tissue entries (sorted by number of experiments) to identify the tissue of interest. Each tissue entry provides basic information about the tissue, along with the number of scRNA-seq experiments associated with it.
(2) Access reference data of specific tissue: Locate the desired tissue, and click the Search button provided within the tissue entry. It will direct you to the Search for Experiment page to access reference data. See section "1.2 Search for annotated reference data" for details.